When imagining the future of technology, sometimes all we need to do is look out the window — or into a microscope.
Our researchers take inspiration from nature to redefine what a computer can be, from data storage using synthetic DNA, to sensors modeled on insects and leaves. We also advance technologies to help solve biology’s biggest mysteries, such as computational approaches for understanding the mechanisms of disease and brain-computer interfaces that can restore or augment physical function and mobility.
Research Groups & Labs
AIMS Lab
The AI for bioMedical Sciences (AIMS) Lab fundamentally advances the way AI is integrated with biology and clinical medicine by addressing novel scientific questions spanning explainable AI, model auditing, disease drivers, and more.
Sensor Systems Laboratory
The Sensor Systems Laboratory invents new sensor systems, devises new ways to power and communicate with them, and develops algorithms for using them, with applications in the domains of bioelectronics, robotics, and ubiquitous computing.
Faculty Members
Adjunct Faculty
Faculty
Adjunct Faculty
Adjunct Faculty
Centers & Initiatives

Institute for Medical Data Science (IMDS)
The Institute forMedical Data Science (IMDS) is a joint effort among the Schools of Medicine and Public Health and the College of Engineering, including the Allen School to lead the development and implementation of cutting-edge AI and data science methods in medical data science. By harnessing the power of AI across diverse health determinants, IMDS aims to improve patient health, provider satisfaction, and healthcare operations, particularly in the Pacific Northwest region.

Center for Neurotechnology (CNT)
The Center for Neurotechnology (CNT) got its start in 2011 as one of several Engineering Research Centers (ERCs) funded by the National Science Foundation. CNT is headquartered at the University of Washington, with core partners at the Massachusetts Institute of Technology and San Diego State University. CNT researchers focus on developing and applying principles of engineered neuroplasticity to revolutionize the treatment of spinal cord injury, stroke and other debilitating neurological conditions.
Highlights
Allen School News

Researchers in the UbiComp Lab and UW Medicine earned an IMWUT Distinguished Paper Award for their work on an app that turns a smartphone into a thermometer.
UW News

In this Q&A, Allen School professor Sheng Wang talks about his work on a new medical AI model, BiomedParse, that works across nine different types of medical images to better predict systemic diseases. Clinicians can load images into the system and ask questions in plain English.
Allen School News
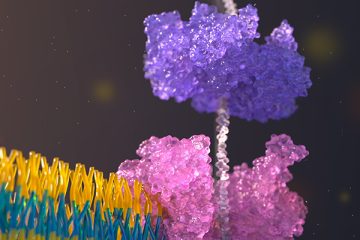
In a paper published in the journal Nature, a team of researchers in the Molecular Information Systems Lab introduced a new approach to long-range, single-molecule protein sequencing by demonstrating how to read each protein molecule by pulling it through a nanopore sensor.




